Using the TimeSeriesAnalysis Package
Anthony G. Sena
2023-08-29
Source:vignettes/UsingTimeSeriesAnalysisPackage.Rmd
UsingTimeSeriesAnalysisPackage.RmdOverview
This document aims to showcase how to use the TimeSeriesAnalysis package for the sample data sets included in the package.
Data set 1: Changes in use of HCQ in RA patients over time
Background: RA guidelines recommend using Methotrexate as first line therapy ASAP from 2008, possibly ‘displacing’ hydroxychloroquine (HCQ).
- Period: 2000 – 2018
outputFolder <- "E:/Timeseries/HCQ_In_RA_Example"
if (dir.exists(outputFolder)) {
unlink(outputFolder, recursive = TRUE)
}
data(drugData)
# HCQ Use in RA Patients is cohortDefinitionId == 3
tsData <- drugData %>%
filter(cohortDefinitionId == 3) %>%
arrange(cohortStartDate) %>%
select(cohortStartDate, subjectCount) %>%
rename(
eventDate = cohortStartDate,
eventCount = subjectCount
)
tsData## # A tibble: 18 x 2
## eventDate eventCount
## <date> <dbl>
## 1 2000-01-01 15
## 2 2001-01-01 21
## 3 2002-01-01 27
## 4 2003-01-01 38
## 5 2004-01-01 37
## 6 2005-01-01 44
## 7 2006-01-01 55
## 8 2007-01-01 62
## 9 2008-01-01 59
## 10 2009-01-01 99
## 11 2010-01-01 109
## 12 2011-01-01 115
## 13 2012-01-01 156
## 14 2013-01-01 142
## 15 2014-01-01 107
## 16 2015-01-01 118
## 17 2016-01-01 102
## 18 2017-01-01 65Creating the model arguments
The following sections will describe how to define the various model fitting arguments for several use cases.
Single Pre-specified change point – fixed (i.e. no estimated changepoint produced by model)
In this example, we’ll create the arguments used to fit a model that has no estimated change points and instead provides a single pre-specified change point for evaluation.
segArgs1 <- createSegmentedArgs(
modelType = "linear",
psi = lubridate::as_date("2001-01-01"),
control = segmented::seg.control(
it.max = 0,
K = 1
)
)Single prespecified changepoint – not fixed (i.e. an estimated changepoint produced by model which may or may no coincide with pre-specified change point)
Now we’ll create the arguments used to fit a model that has 1 estimated change points and a single pre-specified change point for evaluation.
segArgs2 <- createSegmentedArgs(
modelType = "linear",
npsi = 1,
fixed.psi = list(eventDate = lubridate::as_date("2001-01-01"))
)No prespecified changepoint – an estimated changepoint produced by model
Now we’ll create the arguments used to fit a model with a single estimated change point
segArgs3 <- createSegmentedArgs(modelType = "linear")Single prespecified changepoint – and multiple estimated changepoints produced by model
Now we’ll create the arguments used to fit a model with a single pre-specified changepoint and 2 estimated changepoints.
segArgs4 <- createSegmentedArgs(
modelType = "linear",
npsi = 2,
fixed.psi = list(eventDate = lubridate::as_date("2001-01-01"))
)Bayesian Online Change point detection
ocpArgs <- TimeSeriesAnalysis::createOcpArgs()Fitting the models
Next we’ll provide some code for using the arguments and data above to fit the models.
# Create the full set of analyses
tsAnalysis1 <- createTsAnalysis(
analysisId = 1,
description = "Single fixed, pre-specified change point",
tsArgs = segArgs1
)
tsAnalysis2 <- createTsAnalysis(
analysisId = 2,
description = "1 pre-specified, 1 estimated changepoint",
tsArgs = segArgs2
)
tsAnalysis3 <- createTsAnalysis(
analysisId = 3,
description = "Single estimated changepoint",
tsArgs = segArgs3
)
tsAnalysis4 <- createTsAnalysis(
analysisId = 4,
description = "1 pre-specified, 2 estimated changepoints",
tsArgs = segArgs4
)
tsAnalysis5 <- createTsAnalysis(
analysisId = 5,
description = "Bayesian Online Change point detection",
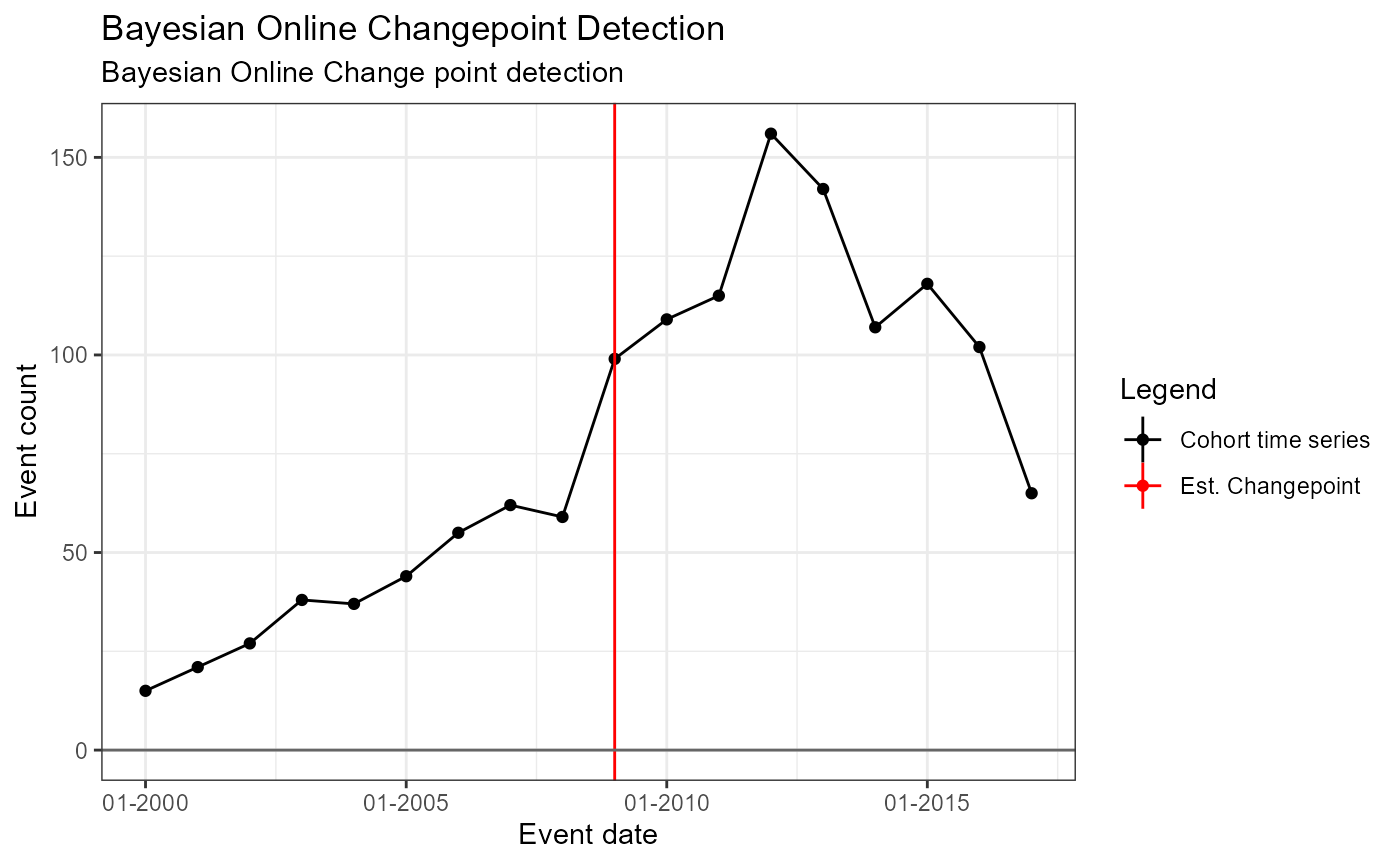
tsArgs = ocpArgs
)
tsAnalysisList <- list(tsAnalysis1, tsAnalysis2, tsAnalysis3, tsAnalysis4, tsAnalysis5)
# Run the analysis
runTsAnalyses(
tsData = tsData,
tsDataId = 1, # A unique identifier for the data set
outputFolder = outputFolder,
tsAnalysisList = tsAnalysisList
)## Building time series models## Analysis 1: Single fixed, pre-specified change point## Analysis 2: 1 pre-specified, 1 estimated changepoint## Analysis 3: Single estimated changepoint## Analysis 4: 1 pre-specified, 2 estimated changepoints## Analysis 5: Bayesian Online Change point detectionInspecting the results
Next we can use the package to plot the original time series and the model estimated change points.
Single Pre-specified change point – fixed (i.e. no estimated changepoint produced by model)
m1 <- readRDS(file = file.path(outputFolder, "Analysis1/ts_d1.rds"))
TimeSeriesAnalysis::plotSegmented(m1$tsData, m1$model, plotSubtitle = tsAnalysisList[[1]]$description)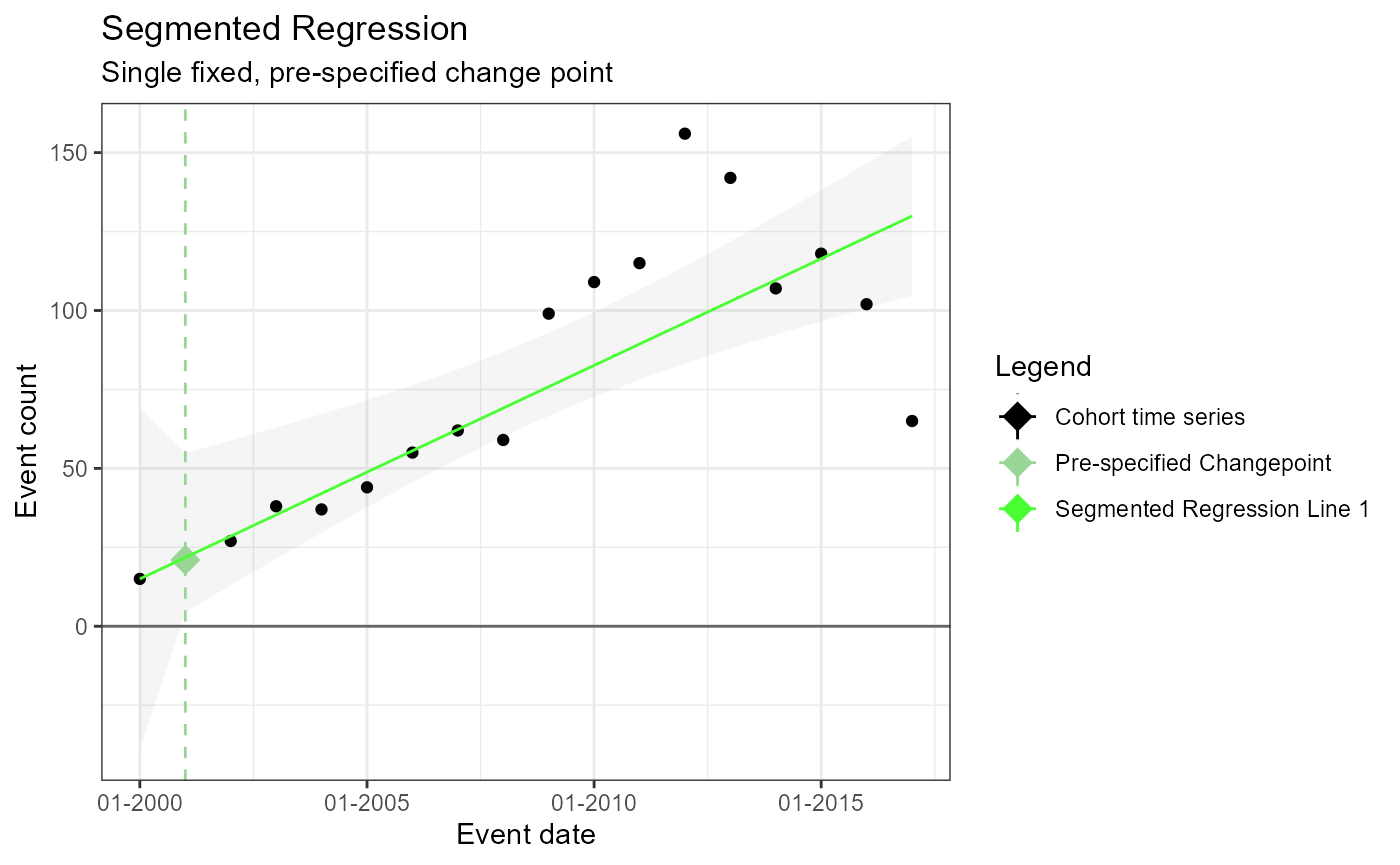
Single prespecified changepoint – not fixed (i.e. an estimated changepoint produced by model which may or may no coincide with pre-specified change point)
m2 <- readRDS(file = file.path(outputFolder, "Analysis2/ts_d1.rds"))
TimeSeriesAnalysis::plotSegmented(m2$tsData, m2$model, plotSubtitle = tsAnalysisList[[2]]$description)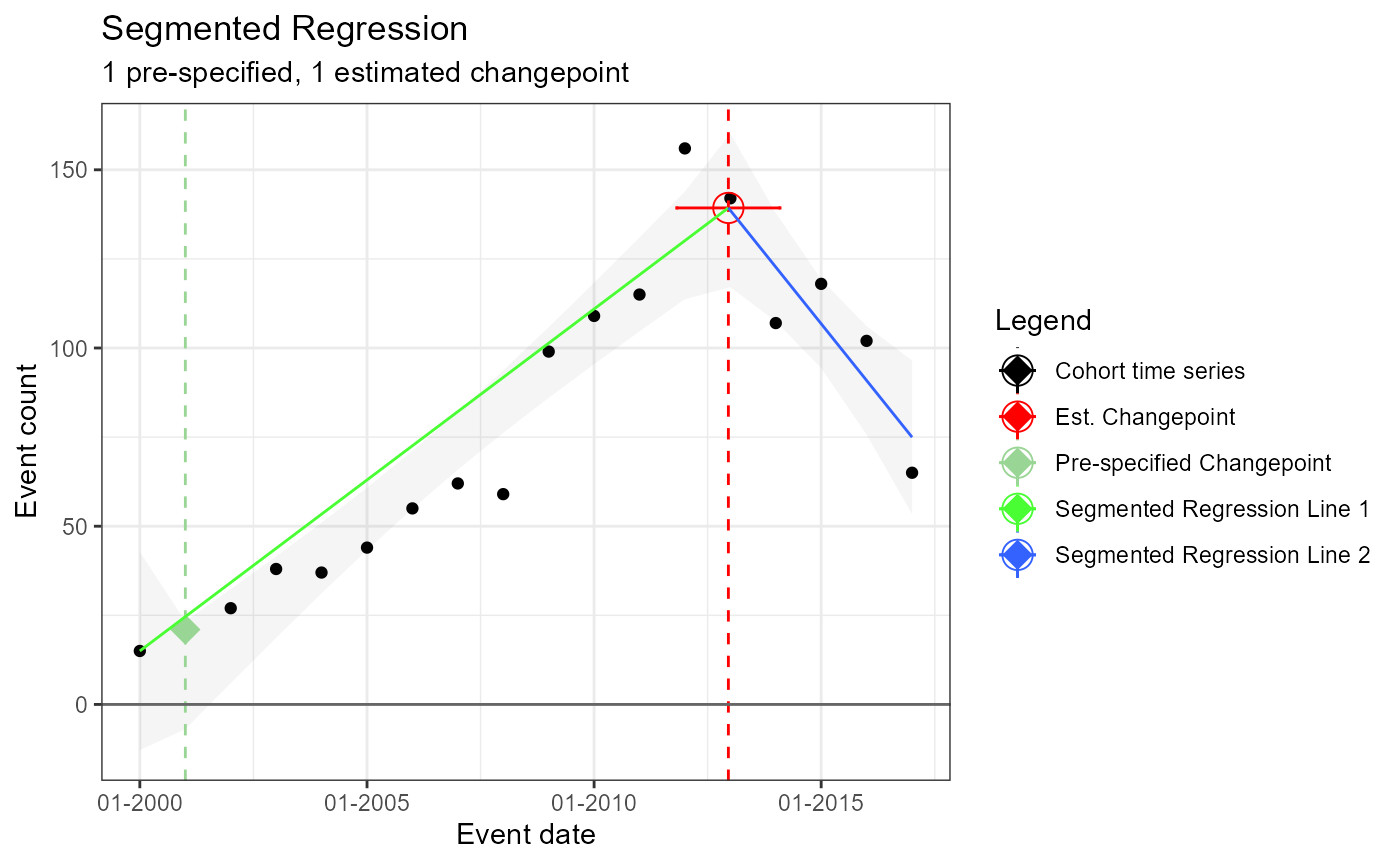
No prespecified changepoint – an estimated changepoint produced by model
m3 <- readRDS(file = file.path(outputFolder, "Analysis3/ts_d1.rds"))
TimeSeriesAnalysis::plotSegmented(m3$tsData, m3$model, plotSubtitle = tsAnalysisList[[3]]$description)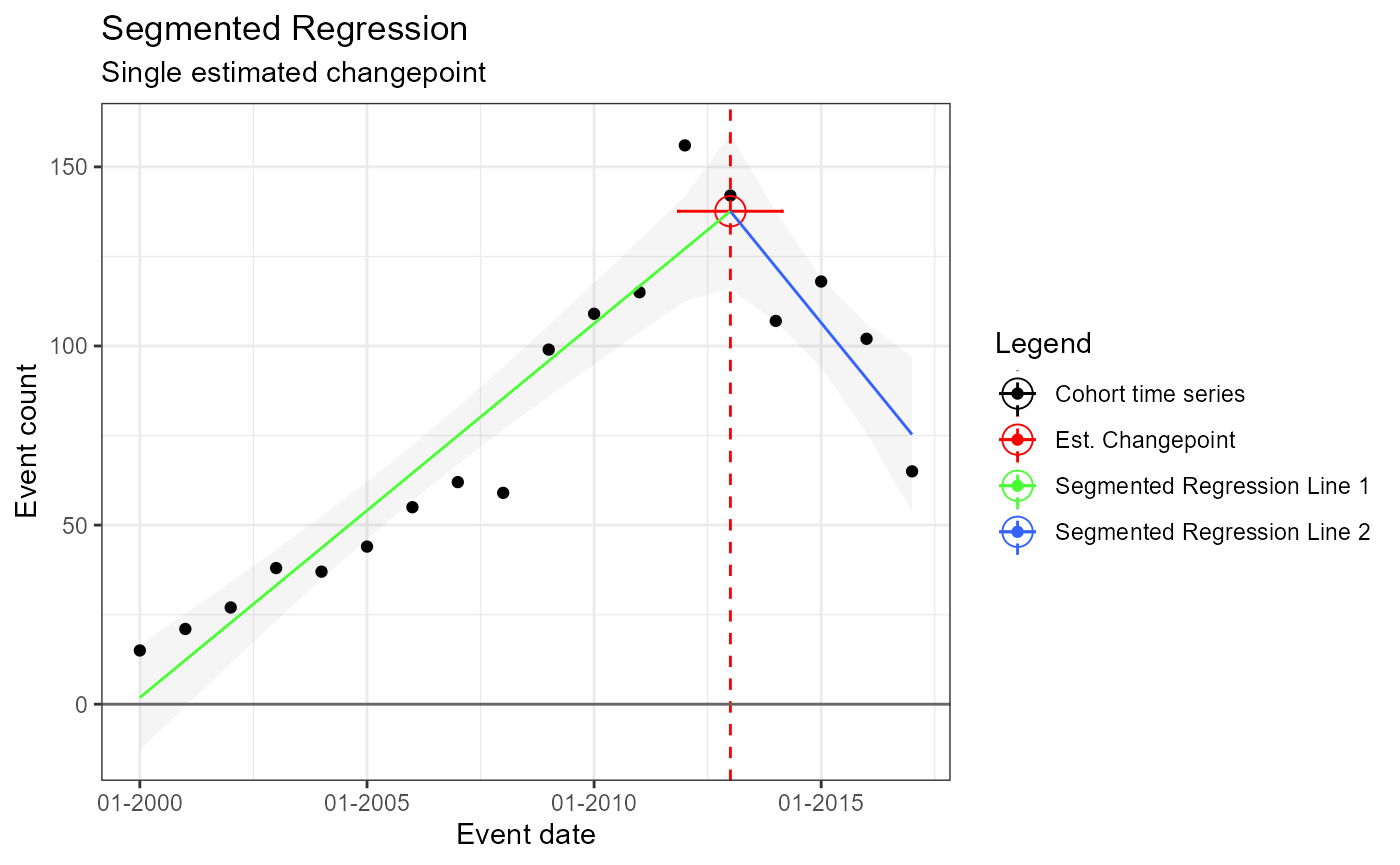
Single prespecified changepoint – and multiple estimated changepoints produced by model
m4 <- readRDS(file = file.path(outputFolder, "Analysis4/ts_d1.rds"))
TimeSeriesAnalysis::plotSegmented(m4$tsData, m4$model, plotSubtitle = tsAnalysisList[[4]]$description)## Warning in matrix(newZ, nrow = nrow(newZ), ncol = length(psi.noti)): data
## length differs from size of matrix: [36 != 18 x 1]
## Warning in matrix(newZ, nrow = nrow(newZ), ncol = length(psi.noti)): data
## length differs from size of matrix: [36 != 18 x 1]## Warning in matrix(newZ, nrow = nrow(newZ), ncol = length(psi.noti)): data
## length differs from size of matrix: [2 != 1 x 1]
## Warning in matrix(newZ, nrow = nrow(newZ), ncol = length(psi.noti)): data
## length differs from size of matrix: [2 != 1 x 1]
## Warning in matrix(newZ, nrow = nrow(newZ), ncol = length(psi.noti)): data
## length differs from size of matrix: [2 != 1 x 1]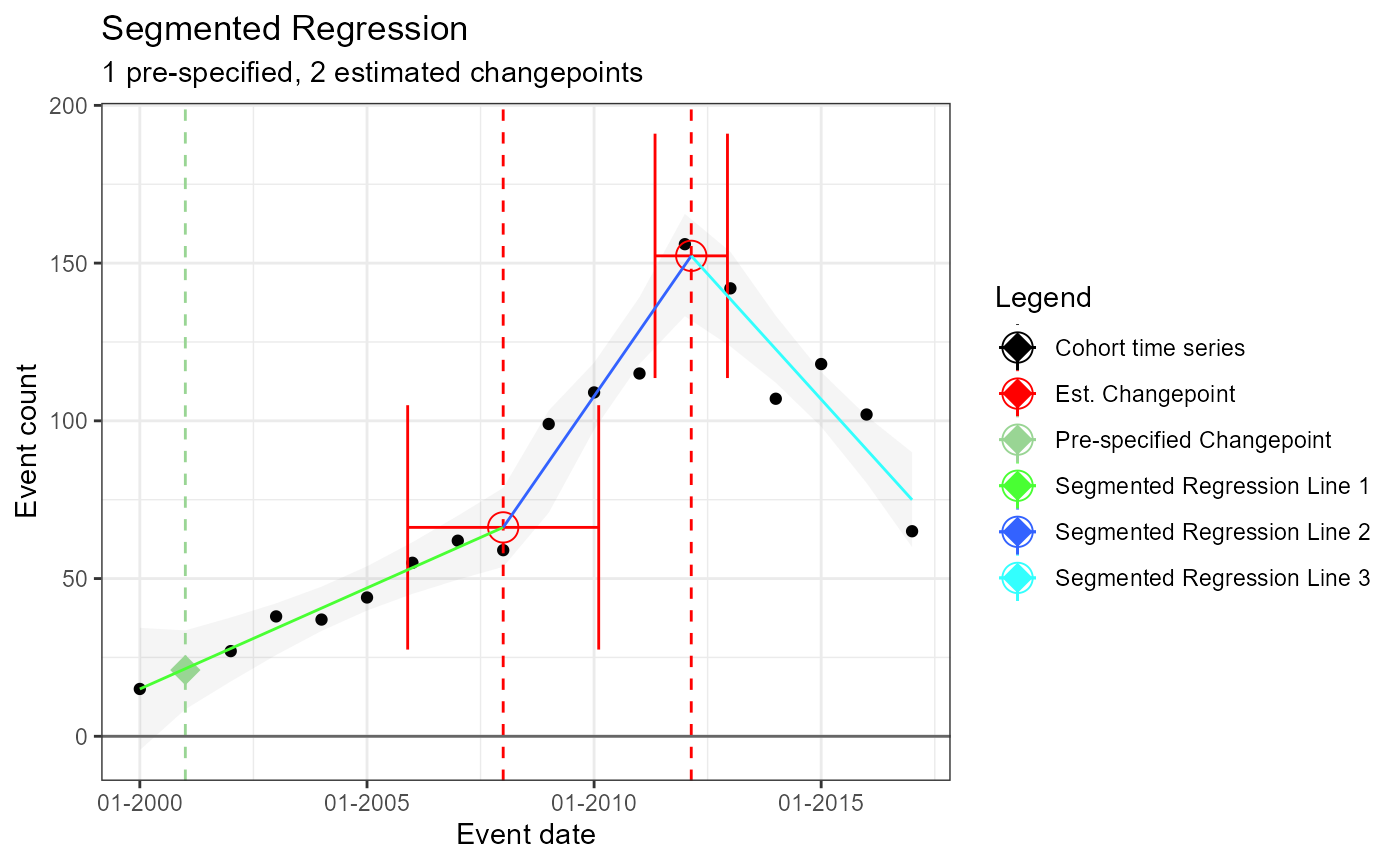
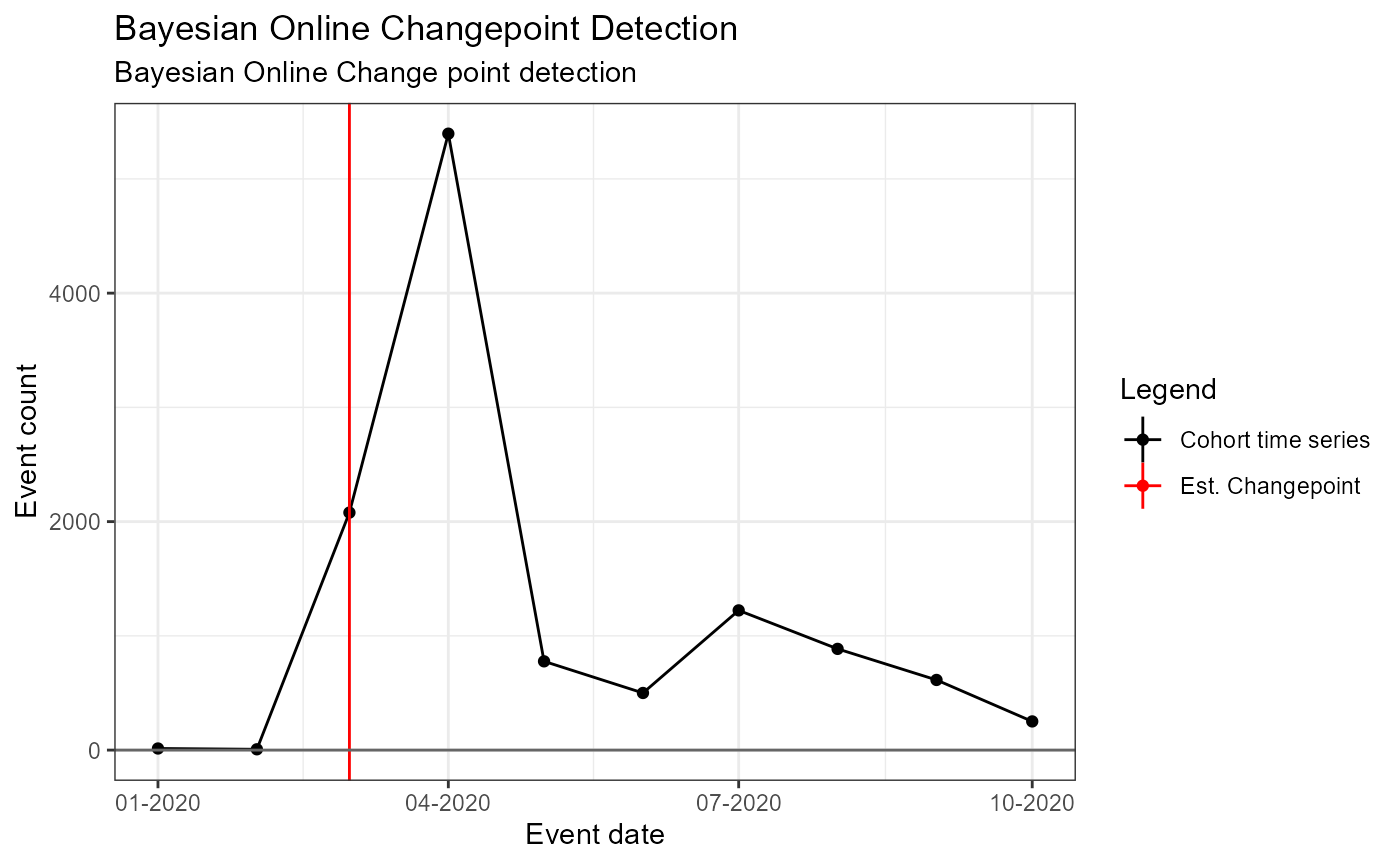
Data Set 2: HCQ for Covid-19 in 2020
Background: - HCQ received emergency approval by FDA in March - Regulatory action followed in late April due to CV safety issues - Data: HCQ for Covid-19 treatment - Period: Jan – Oct 2020
outputFolder <- "E:/Timeseries/HCQ_In_COVID"
if (dir.exists(outputFolder)) {
unlink(outputFolder, recursive = TRUE)
}
# HCQ for Covid-19 in 2020 is cohortDefinitionId == 1
tsData <- drugData %>%
filter(cohortDefinitionId == 1) %>%
arrange(cohortStartDate) %>%
select(cohortStartDate, subjectCount) %>%
rename(
eventDate = cohortStartDate,
eventCount = subjectCount
)
tsData## # A tibble: 10 x 2
## eventDate eventCount
## <date> <dbl>
## 1 2020-01-01 14
## 2 2020-02-01 7
## 3 2020-03-01 2079
## 4 2020-04-01 5396
## 5 2020-05-01 777
## 6 2020-06-01 500
## 7 2020-07-01 1223
## 8 2020-08-01 886
## 9 2020-09-01 614
## 10 2020-10-01 251Single Pre-specified change point – fixed (i.e. no estimated changepoint produced by model)
segArgs1 <- createSegmentedArgs(
modelType = "linear",
psi = lubridate::as_date("2020-03-01"),
control = segmented::seg.control(
it.max = 0,
K = 1
)
)Single prespecified changepoint – not fixed (i.e. an estimated changepoint produced by model which may or may no coincide with pre-specified change point)
segArgs2 <- createSegmentedArgs(
modelType = "linear",
npsi = 1,
fixed.psi = list(eventDate = lubridate::as_date("2020-03-01"))
)No prespecified changepoint – an estimated changepoint produced by model
Now we’ll create the arguments used to fit a model with a single estimated change point
segArgs3 <- createSegmentedArgs(modelType = "linear")Single prespecified changepoint – and multiple estimated changepoints produced by model
Now we’ll create the arguments used to fit a model with a single pre-specified changepoint and 2 estimated changepoints.
segArgs4 <- createSegmentedArgs(
modelType = "linear",
npsi = 2,
fixed.psi = list(eventDate = lubridate::as_date("2020-03-01"))
)Fitting the models
Next we’ll provide some code for using the arguments and data above to fit the models.
# Create the full set of analyses
tsAnalysis1 <- createTsAnalysis(
analysisId = 1,
description = "Single fixed, pre-specified change point",
tsArgs = segArgs1
)
tsAnalysis2 <- createTsAnalysis(
analysisId = 2,
description = "1 pre-specified, 1 estimated changepoint",
tsArgs = segArgs2
)
tsAnalysis3 <- createTsAnalysis(
analysisId = 3,
description = "Single estimated changepoint",
tsArgs = segArgs3
)
tsAnalysis4 <- createTsAnalysis(
analysisId = 4,
description = "1 pre-specified, 2 estimated changepoints",
tsArgs = segArgs4
)
tsAnalysis5 <- createTsAnalysis(
analysisId = 5,
description = "Bayesian Online Change point detection",
tsArgs = ocpArgs
)
tsAnalysisList <- list(tsAnalysis1, tsAnalysis2, tsAnalysis3, tsAnalysis4, tsAnalysis5)
# Run the analysis
runTsAnalyses(
tsData = tsData,
tsDataId = 2, # A unique identifier for the data set
outputFolder = outputFolder,
tsAnalysisList = tsAnalysisList
)## Building time series models## Analysis 1: Single fixed, pre-specified change point## Analysis 2: 1 pre-specified, 1 estimated changepoint## Warning encountered when fitting segmented model: No breakpoint estimated## Analysis 3: Single estimated changepoint## Analysis 4: 1 pre-specified, 2 estimated changepoints## breakpoint estimate(s): 18353 18444## Error encountered when fitting segmented model: at least one coef is NA: breakpoint(s) at the boundary? (possibly with many x-values replicated)## Analysis 5: Bayesian Online Change point detectionSingle Pre-specified change point – fixed (i.e. no estimated changepoint produced by model)
m1 <- readRDS(file = file.path(outputFolder, "Analysis1/ts_d2.rds"))
TimeSeriesAnalysis::plotSegmented(m1$tsData, m1$model, plotSubtitle = tsAnalysisList[[1]]$description)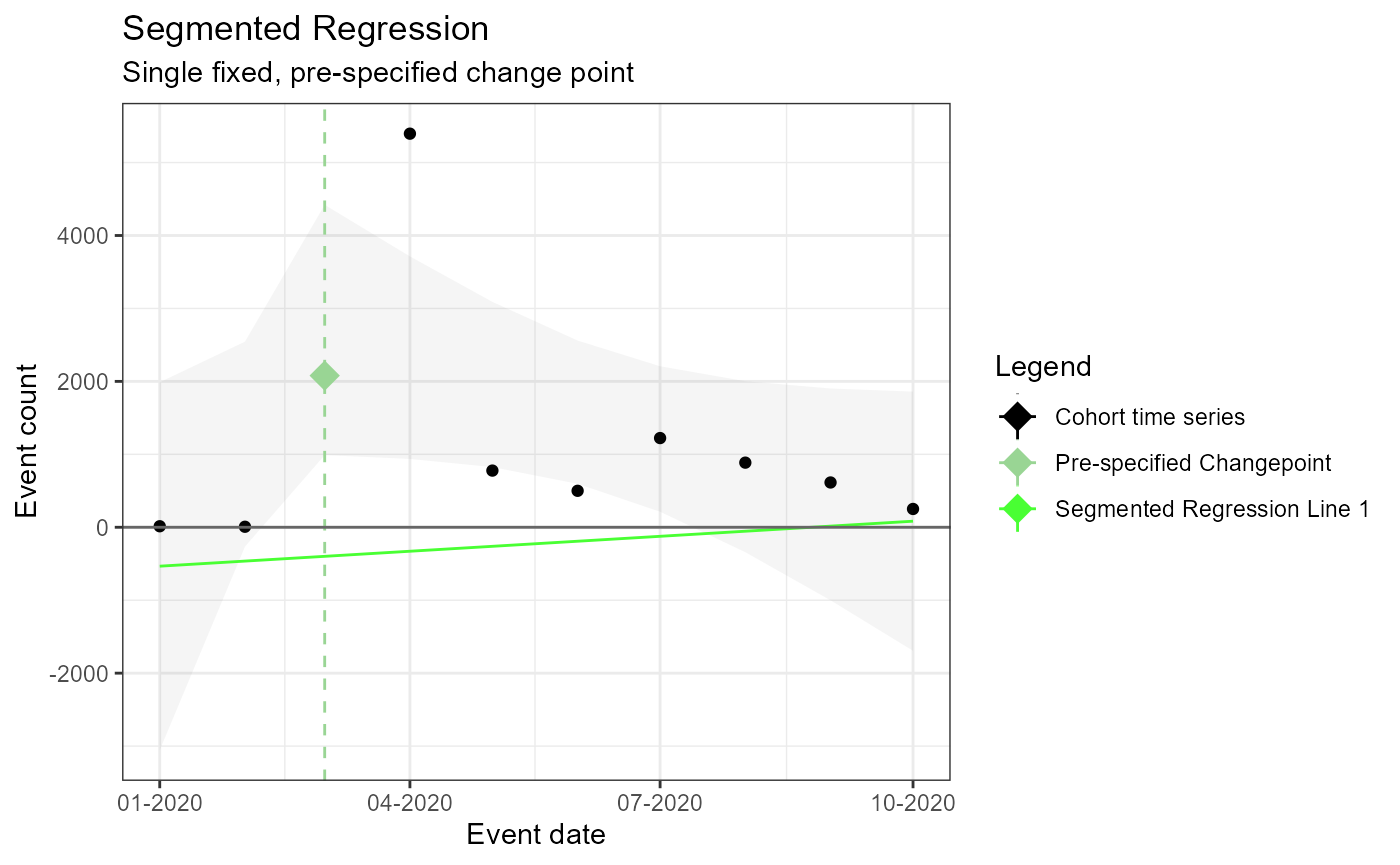
Single prespecified changepoint – not fixed (i.e. an estimated changepoint produced by model which may or may no coincide with pre-specified change point)
m2 <- readRDS(file = file.path(outputFolder, "Analysis2/ts_d2.rds"))
TimeSeriesAnalysis::plotSegmented(m2$tsData, m2$model, plotSubtitle = tsAnalysisList[[2]]$description)## Error in tryCatch(withCallingHandlers({: 1 assertions failed:
## * Variable '!is.null(model)': Must be TRUE.No prespecified changepoint – an estimated changepoint produced by model
m3 <- readRDS(file = file.path(outputFolder, "Analysis3/ts_d2.rds"))
TimeSeriesAnalysis::plotSegmented(m3$tsData, m3$model, plotSubtitle = tsAnalysisList[[3]]$description)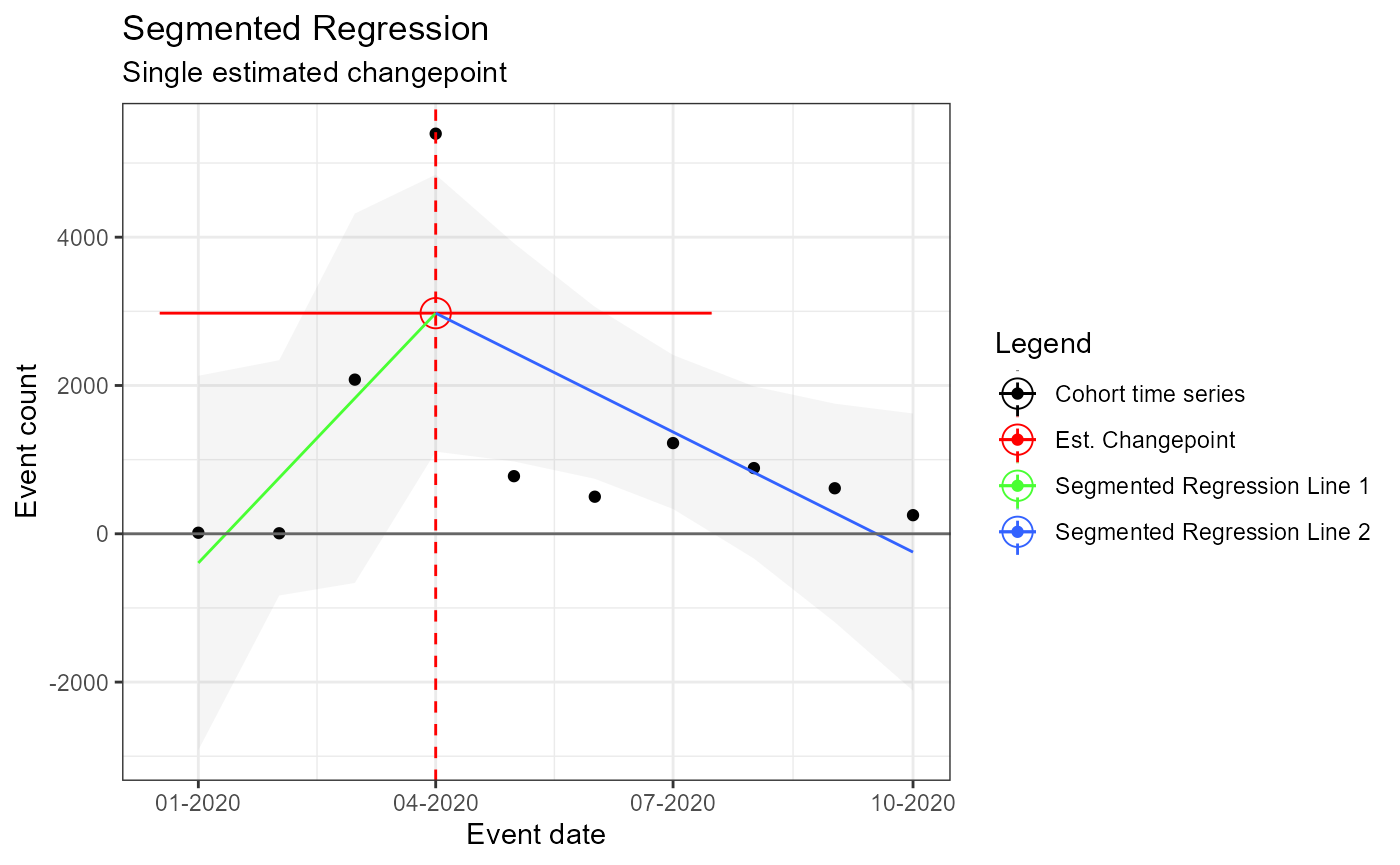
Single prespecified changepoint – and multiple estimated changepoints produced by model
m4 <- readRDS(file = file.path(outputFolder, "Analysis4/ts_d2.rds"))
TimeSeriesAnalysis::plotSegmented(m4$tsData, m4$model, plotSubtitle = tsAnalysisList[[4]]$description)## Error in tryCatch(withCallingHandlers({: 1 assertions failed:
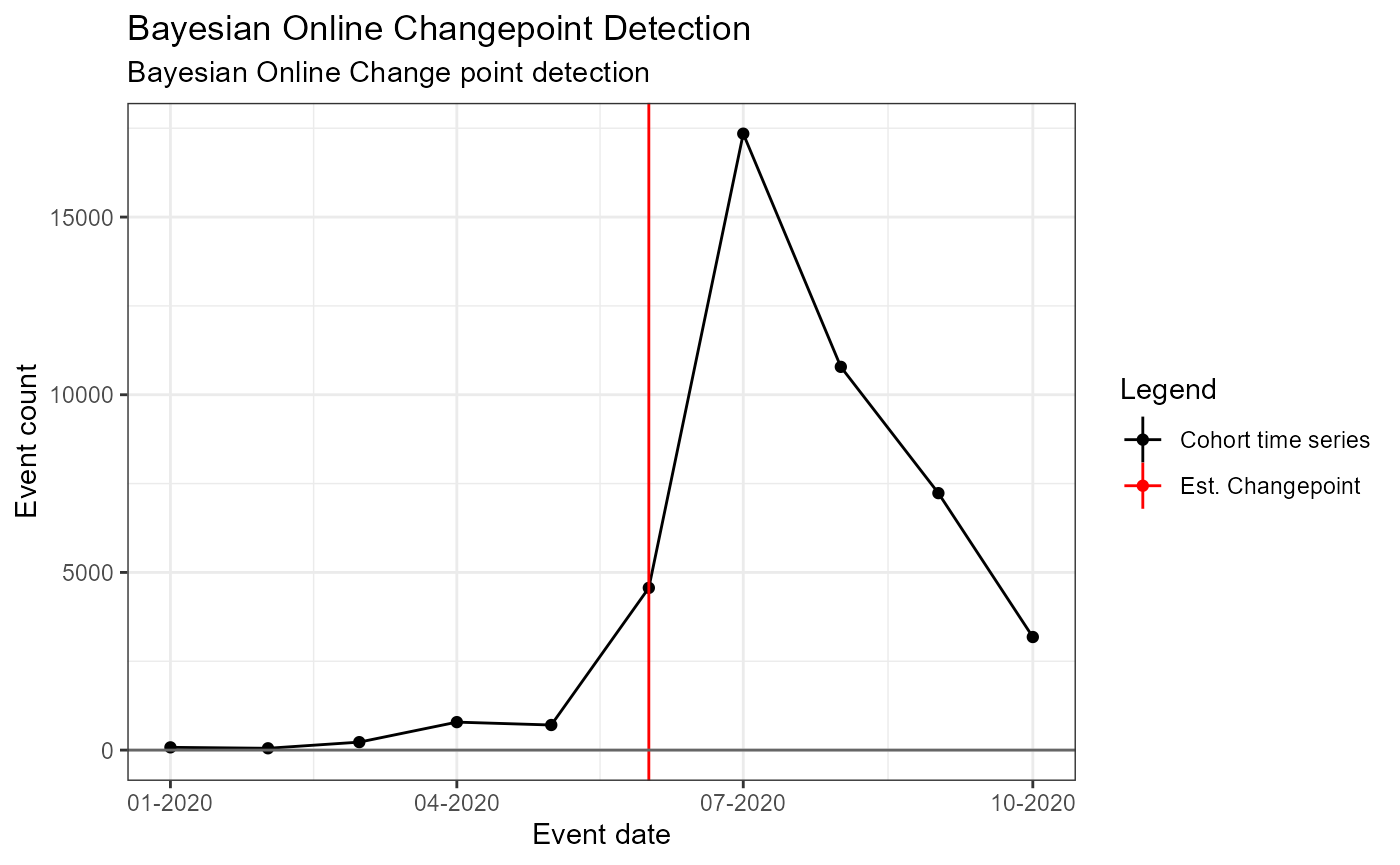
## * Variable '!is.null(model)': Must be TRUE.Data Set 3: Dexamethasone for Covid-19 in 2020
Background: - RECOVERY RCT report 30% reduction in mortality in late June 2020 with dexamethasone (DXM) - Period: Jan – Oct 2020
outputFolder <- "E:/Timeseries/DEX_In_COVID"
if (dir.exists(outputFolder)) {
unlink(outputFolder, recursive = TRUE)
}
# DEX for Covid-19 in 2020 is cohortDefinitionId == 2
tsData <- drugData %>%
filter(cohortDefinitionId == 2) %>%
arrange(cohortStartDate) %>%
select(cohortStartDate, subjectCount) %>%
rename(
eventDate = cohortStartDate,
eventCount = subjectCount
)
tsData## # A tibble: 10 x 2
## eventDate eventCount
## <date> <dbl>
## 1 2020-01-01 77
## 2 2020-02-01 51
## 3 2020-03-01 224
## 4 2020-04-01 787
## 5 2020-05-01 706
## 6 2020-06-01 4565
## 7 2020-07-01 17347
## 8 2020-08-01 10785
## 9 2020-09-01 7231
## 10 2020-10-01 3182Single Pre-specified change point – fixed (i.e. no estimated changepoint produced by model)
segArgs1 <- createSegmentedArgs(
modelType = "linear",
psi = lubridate::as_date("2020-06-01"),
control = segmented::seg.control(
it.max = 0,
K = 1
)
)Single prespecified changepoint – not fixed (i.e. an estimated changepoint produced by model which may or may no coincide with pre-specified change point)
segArgs2 <- createSegmentedArgs(
modelType = "linear",
npsi = 1,
fixed.psi = list(eventDate = lubridate::as_date("2020-06-01"))
)No prespecified changepoint – an estimated changepoint produced by model
Now we’ll create the arguments used to fit a model with a single estimated change point
segArgs3 <- createSegmentedArgs(modelType = "linear")Single prespecified changepoint – and multiple estimated changepoints produced by model
Now we’ll create the arguments used to fit a model with a single pre-specified changepoint and 2 estimated changepoints.
segArgs4 <- createSegmentedArgs(
modelType = "linear",
npsi = 2,
fixed.psi = list(eventDate = lubridate::as_date("2020-06-01"))
)Fitting the models
Next we’ll provide some code for using the arguments and data above to fit the models.
# Create the full set of analyses
tsAnalysis1 <- createTsAnalysis(
analysisId = 1,
description = "Single fixed, pre-specified change point",
tsArgs = segArgs1
)
tsAnalysis2 <- createTsAnalysis(
analysisId = 2,
description = "1 pre-specified, 1 estimated changepoint",
tsArgs = segArgs2
)
tsAnalysis3 <- createTsAnalysis(
analysisId = 3,
description = "Single estimated changepoint",
tsArgs = segArgs3
)
tsAnalysis4 <- createTsAnalysis(
analysisId = 4,
description = "1 pre-specified, 2 estimated changepoints",
tsArgs = segArgs4
)
tsAnalysis5 <- createTsAnalysis(
analysisId = 5,
description = "Bayesian Online Change point detection",
tsArgs = ocpArgs
)
tsAnalysisList <- list(tsAnalysis1, tsAnalysis2, tsAnalysis3, tsAnalysis4, tsAnalysis5)
# Run the analysis
runTsAnalyses(
tsData = tsData,
tsDataId = 3, # A unique identifier for the data set
outputFolder = outputFolder,
tsAnalysisList = tsAnalysisList
)## Building time series models## Analysis 1: Single fixed, pre-specified change point## Analysis 2: 1 pre-specified, 1 estimated changepoint## Warning encountered when fitting segmented model: No breakpoint estimated## Analysis 3: Single estimated changepoint## Analysis 4: 1 pre-specified, 2 estimated changepoints## Warning encountered when fitting segmented model: No breakpoint estimated## Analysis 5: Bayesian Online Change point detectionSingle Pre-specified change point – fixed (i.e. no estimated changepoint produced by model)
m1 <- readRDS(file = file.path(outputFolder, "Analysis1/ts_d3.rds"))
TimeSeriesAnalysis::plotSegmented(m1$tsData, m1$model, plotSubtitle = tsAnalysisList[[1]]$description)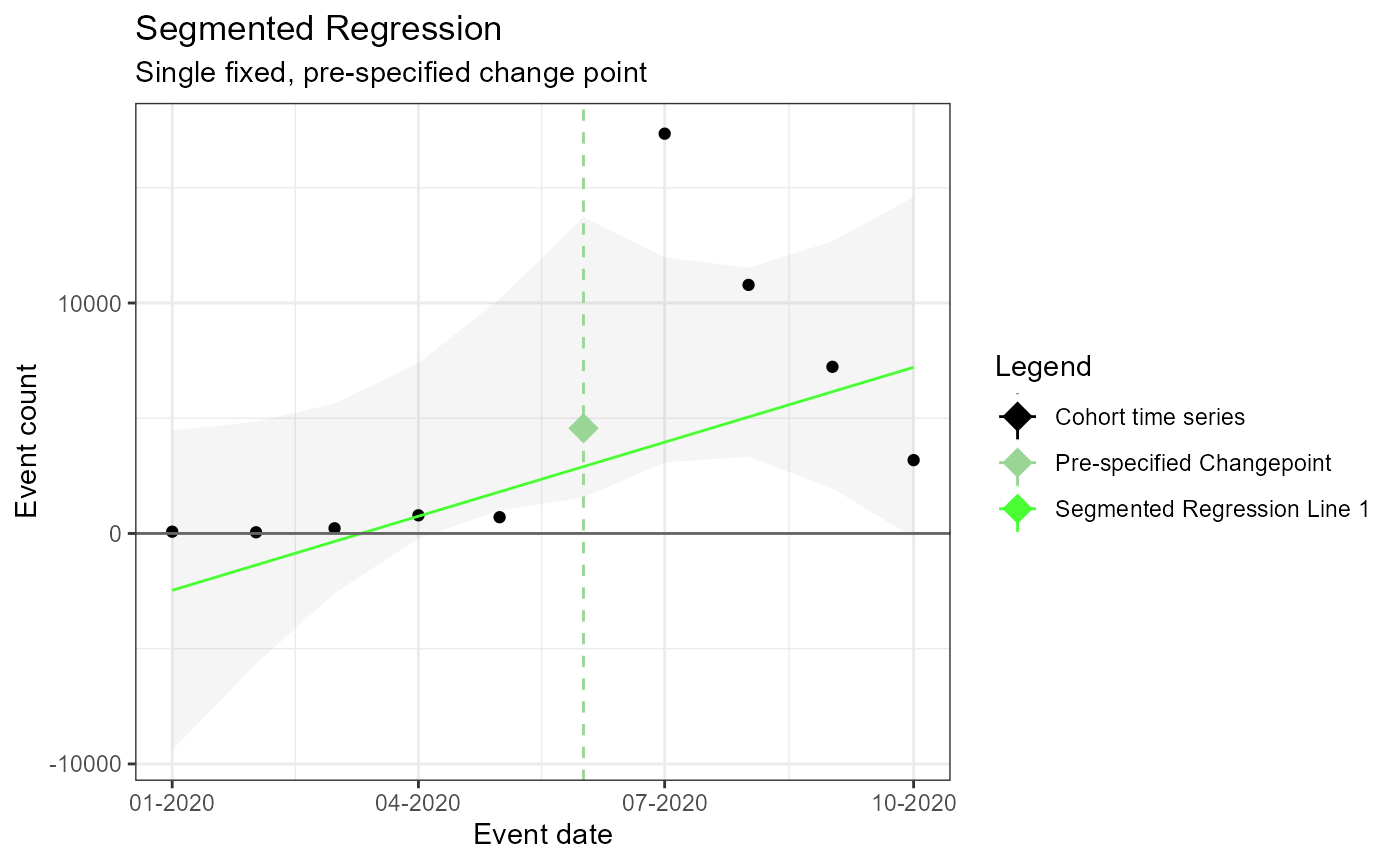
Single prespecified changepoint – not fixed (i.e. an estimated changepoint produced by model which may or may no coincide with pre-specified change point)
m2 <- readRDS(file = file.path(outputFolder, "Analysis2/ts_d3.rds"))
TimeSeriesAnalysis::plotSegmented(m2$tsData, m2$model, plotSubtitle = tsAnalysisList[[2]]$description)## Error in tryCatch(withCallingHandlers({: 1 assertions failed:
## * Variable '!is.null(model)': Must be TRUE.No prespecified changepoint – an estimated changepoint produced by model
m3 <- readRDS(file = file.path(outputFolder, "Analysis3/ts_d3.rds"))
TimeSeriesAnalysis::plotSegmented(m3$tsData, m3$model, plotSubtitle = tsAnalysisList[[3]]$description)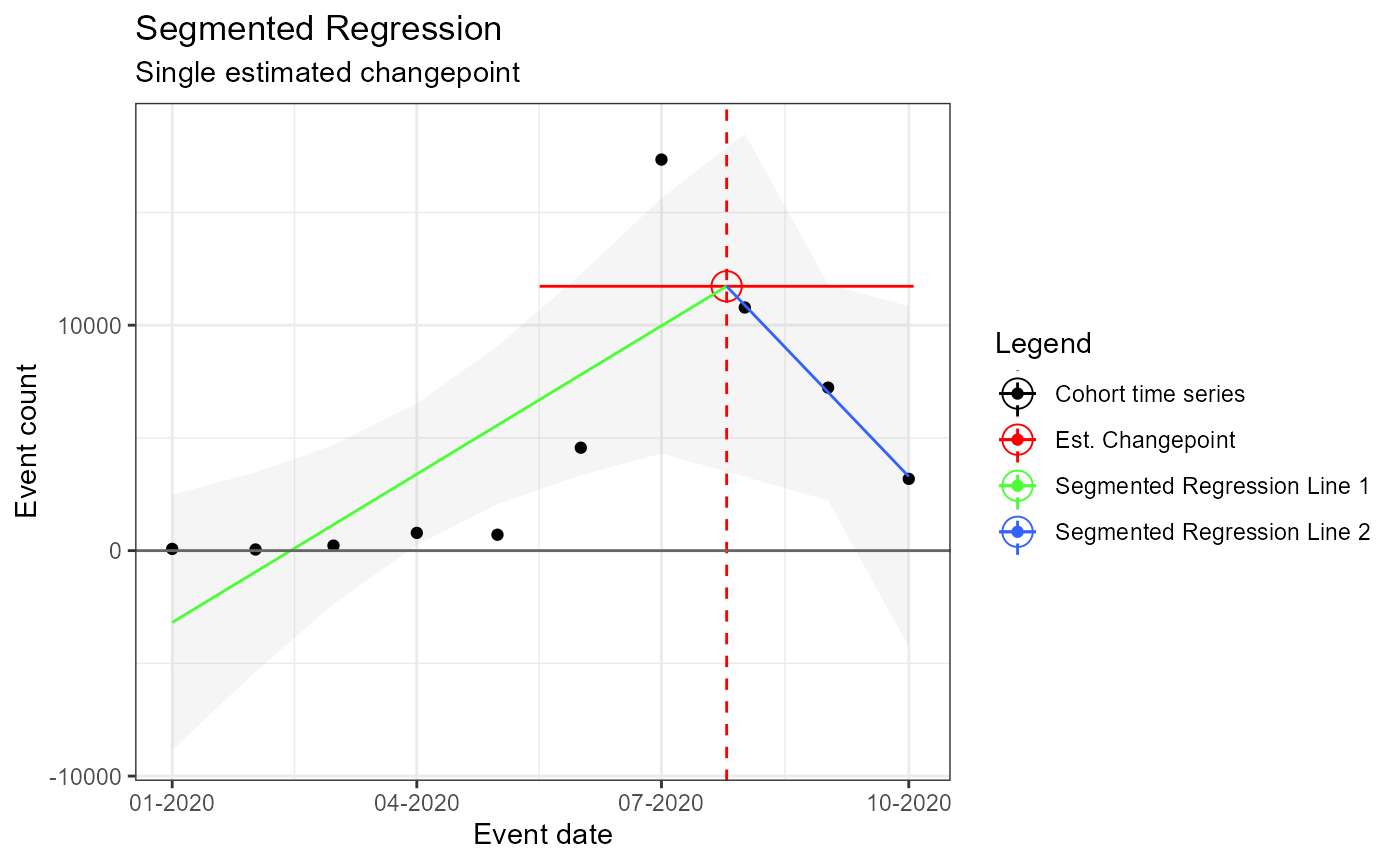
Single prespecified changepoint – and multiple estimated changepoints produced by model
m4 <- readRDS(file = file.path(outputFolder, "Analysis4/ts_d3.rds"))
TimeSeriesAnalysis::plotSegmented(m4$tsData, m4$model, plotSubtitle = tsAnalysisList[[4]]$description)## Error in tryCatch(withCallingHandlers({: 1 assertions failed:
## * Variable '!is.null(model)': Must be TRUE.