UK BioBank to OMOP CDM v5.3.1
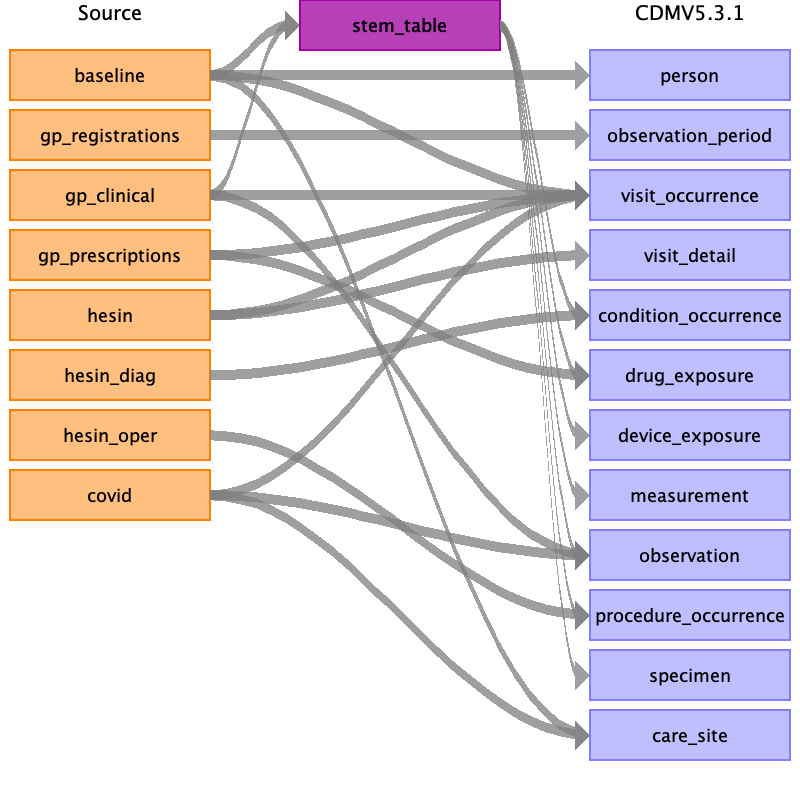
The UK BioBank data consists of five sections
- Baseline, survey and lab results collected during visits to the assessment centres
- Hospital Episode Statistic, inpatient care
- General Practitioner, outpatient care
- Covid registry
- Death registry
Here we specify the mapping of each of these sections to the respective OMOP tables.
Health System data
Care_site
Clinical data
Person
Observation_period
Death
Visit_occurrence
We use a heuristic to ‘calculate’ a unique 16 digit visit_occurrence_id for each of the sources. The id is a concatenation of a source digit (1 digit), the eid (7 digits) and an index unique within the patient (filled to 8 digits). What field is used as the index is specific to the source table, see overview below. Note that the visit_occurence_id has to be a Big Integer to be able to hold 16 digits.
From baseline: 1<eid><instance>, e.g. 1_9876543_00000001
From covid: 2<eid><date>, e.g. 2_9876543_20201231
From hesin: 3<eid><spell_index>, e.g. 3_9876543_00000012
From gp_clinical and gp_prescriptions: 4<eid><date>, e.g. 4_9876543_20201231
Visit_detail
We use a heuristic to ‘calculate’ a unique 11 digit visit_detail_id. As the hesin table is the only source for visit details, we only need to concatenate the eid and an index (the ins_index).
From hesin: <eid><ins_index>, e.g. 9876543_0084
Condition_occurrence
Drug_exposure
Procedure_occurrence
Observation
Stem_table
Stem_to_clinical_event

The stem table is mapped to the respective OMOP domains based on the domain_id. The following rules are applied, in this order:
- If
stem_table.domain_idgiven, then read target domain fromstem_table.domain_id - If
stem_table.concept_idnot 0, then read target domain fromconcept.domain_id - Else, the target domain is Observation.
- To condition_occurrence
- To drug_exposure
- To procedure_occurrence
- To device_exposure
- To measurement
- To specimen